FAIR Annotation of Bioassay Metadata – Pistoia Alliance
The Pistoia Alliance Bioassay FAIR Annotation project develops digital standards for bioassay metadata, provides annotations of bioassay method descriptions according to these standards, and makes them available publicly.
- Proposed a minimal information model for assay metadata. It is now used by the FDA IVP project.
- Annotated close to 2,800 previously published assay methods.
Overview
Most of the information in modern biological research comes from standardised assays. Descriptions of assays and assay methods constitute the assay metadata for the measurements produced by these assays. It has been estimated that over a million unstructured bioassay descriptions are available in the public domain (see PubChem Statistics), and additional ones are being continuously published. Assay method descriptions are extremely important for research planning, for selection of methods, equipment and reagents, for regulatory submissions, and for avoidance of experiment approaches known to have failed in the past. An ability to quickly find and interpret assays and assay metadata is therefore central to the quality of the pharmaceutical R&D process. Further, when building AI models based on assay data, one must consider metadata as well, or risk missing the appropriate data or inclusion of inappropriate data into the model. Another aspect of creating standardised assay data for AI is the creation of ready-to-use data eliminating the data-cleaning and preprocessing steps for most cases. In addition, these assay method descriptions form a rich source of information for post-hoc data mining on their own.
However, assay protocols have not been available in sufficient level of granularity and machine-readable formats suitable for automated processing or data mining. This project aims to remedy that. We work with the scientific community to develop standards for representation of assay metadata. We convert publicly available plain text assay method descriptions into a structured format that uses the proposed standards. This annotation process makes the assay method descriptions findable, accessible, interoperable, and reusable (FAIR). In an ideal future state, the new assay methods will be published in the digital form directly using the set of standards agreed upon by the scientific community. This approach has the potential to greatly improve the quality and reproducibility of research.
Process
At this point in time, we have completed a pilot project and one production run of assay metadata curation.
In the pilot project, the main objective was to establish the proof-of-principle for assay FAIR metadata annotation:
- We conducted a search for NLP technology that would be suitable for the assay metadata annotation. Collaborative Drug Discovery BioHarmony Annotator was selected in an open request for proposal process. BioHarmony Annotator is convenient to use with a hybrid ML/human-in-the-loop approach; where the initial NLP data extraction and automated annotation suggestions are followed by human expert review.
- We collected key business questions that are frequently addressed in meta-analysis, and defined a data model that allows one to easily address these questions. We relied on the BioAssay Ontology (BAO) and other standard ontologies in formulation of the data model, and communicated the desired changes back to BAO.
- We published this data model in BioHarmony Annotator, open to the general public with minimal registration requirements, and as a CEDAR metadata template (Stanford University https://metadatacenter.org/), and disclosed it in a series of public talks.
- We analysed and annotated 496 assay descriptions from three sources of public bioassay metadata, and placed a subset of these annotated assay descriptions into PubChem. The three sources were: (1) Descriptions of assays targeting EGFR inhibition by small molecules, obtained predominantly from peer-reviewed papers; (2) PubChem assay descriptions selected from HTS datasets generated with the NCATS CANVASS Library; and (3) vendor assay descriptions selected from the Thermo-Fisher Z’-LYTE kinase panel, used with permission. These assays are commonly used in the pharmaceutical industry, and publication of additional metadata should benefit the scientific community while also enriching internal company databases.
- We investigated an operational model in which pharmaceutical industry partners jointly paid for high quality curation of publicly available bioassay descriptions.
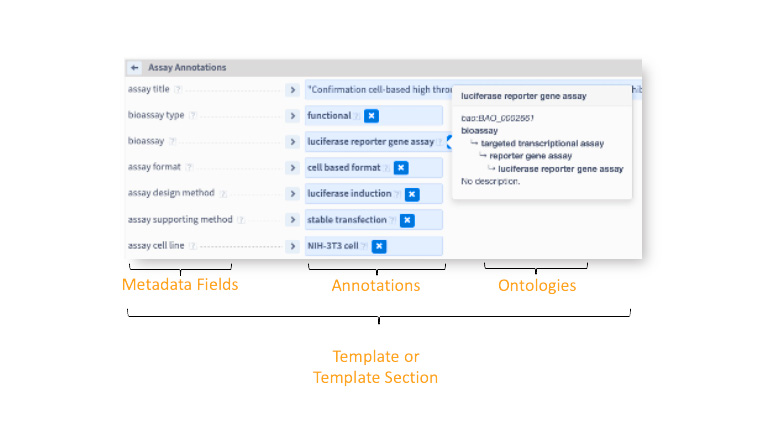
An illustration of the assay metadata curation interface in the BioHarmony Annotator
On the basis of the observations made in the pilot project, we concluded that the vendor assay panel descriptions are the best choice of assay metadata. Consequently, in the first production run of assay metadata annotation, we focused on the assay categories that are associated with commercial providers:
|
Assay category
|
Assay count |
| Pubchem COVID-19 Assays | 624 |
| PubChem Phospholipidosis Assays | 632 |
| KINOME Scan | 489 |
| Eurofins (Cerep) Safety Screen 44 | 44 |
| Star Protocols by Cell Press | 68 |
Efforts to deposit the completed assay annotations into PubChem are currently ongoing. We are also actively engaged in efforts to harmonize our assay metadata annotation template with the proprietary assay registration systems used by the commercial firm members of the Pistoia Alliance and with the new digital In-Vitro Pharmacology initiative by the US FDA. We are also coordinating with the Society for Laboratory Automation and Screening (SLAS) and Elsevier publishing to promote the assay metadata annotation template.
Outcomes
- We successfully created a data model for the assay methods descriptions based on the business needs of the member organisations, and then used this data model to annotate a collection of 2353 assay protocols picked from three sources. Commercial assay panels were the easiest to annotate, while the assay protocols from peer- reviewed papers were the most challenging. To maximise return-of-investment, future annotation efforts should thus be focused on commercially available assay panels.
- We found a number of issues associated with publishing of assay method descriptions that may severely impact the quality and reproducibility of scientific research, and may also have implications for regulatory compliance. In particular, assay methods may contain errors that are propagated from paper-to-paper and are not discovered despite peer review. Also, in some cases, the methods in publications refer to discontinued commercial assay protocols for which full descriptions are no longer available. Last, assay method descriptions for selectivity assays may report less detail than the ones describing the main target of the paper. This may affect the reproducibility of these experiments.
- We established good work practices for the annotation of assay protocols: use human experts to verify the automated annotations, maintain an audit trail and version history, and perform human annotation iteratively. Assessing the AI capabilities of the Bioharmony Annotator and the amount of human expertise needed for this process.
- There is a clear need for a community data standard for future assay publications, that should preferably be made in a digital form (in place of, or in addition to, a traditional plain text publication). We are working with publishers (Elsevier and SLAS), the US FDA, National Institute of Environmental Health Sciences (NIEHS, part of the US NIH), individual Pistoia Alliance member firms, and related Pistoia Alliance projects to define a new industry standard for communication and reporting assay metadata.
Conclusions
We are advancing a project in large-scale annotation of assay protocols using NLP technology with human-in-the-loop quality control. We were successful at establishing the data model, identifying suitable software technologies, identifying data sources for assay protocol descriptions, and annotating over 2300 assay protocols to date. We have aligned with the major academic and government institutions that either publish or use assay metadata (US FDA, PubChem, NIH, BioAssay Ontology, University of Miami, Stanford University, SLAS). In the next phase we plan to increase annotation effort for already published assay protocols, aiming to maximum value for our member organisations, and to promote the emergent data standard for broader adoption by the research community.
References and Resources
- Z-LYTE panel: https://www.thermofisher.com/uk/en/home/industrial/pharma- biopharma/drug-discovery-development/target-and-lead-identification-and- validation/kinasebiology/kinase-activity-assays/z-lyte.html
- BioHarmony Common Assay Template: https://www.bioassayexpress.com/diagnostics/template.jsp#1
- BioAssay Ontology: http://bioassayontology.org
- PubChem: https://pubchem.ncbi.nlm.nih.gov/ and statistics at https://pubchem.ncbi.nlm.nih.gov/docs/statistics
At a Glance
Timeline
- 30 months
Deliverables to-date
- Defined metadata model
- Created a process for automated annotation of assay metadata
- Upload results to a public resource (PubChem)
Authors
- Isabella Feierberg, Jnana Therapeutics (formerly at AstraZeneca)
- Dana Vanderwall, Digital Lab Consulting (formerly at BMS)
- Samantha Jeschonek, PerkinElmer (formerly at Collaborative Drug Discovery)
- Gabriel Backiananthan, Novartis
- Vladimir Makarov, Pistoia Alliance
- Thomas Liener, formerly at Pistoia Alliance
(Many more people were involved in this project. This is just a list of authors of the use case).
Top Tips
- FAIR annotation of assay protocols benefits the scientific community in making research easier to reproduce, and scientific data easier to mine post-hoc; makes the publication of assay methods and protocols easier and improves quality; may reduce work effort in regulatory submissions; and may also help vendors of assay kits and reagents in marketing of their products.
- The best source of assay annotation information is not peer-reviewed papers, that are actually very hard to mine, but vendor-published protocols and instructions pages.
- There is a need for a minimal information standard for assay protocols.
Method tags: BioAssay, Ontologies, PubChem

