FAIRifying Collaborative Research on Real World Data
The Hyve present an approach to making new collaborative scientific research data FAIR in a real time manner on the internet.
- Rapid development of a semantic model expressed as subset of schema.org
- Reusable static web site generator code

Overview
The IMI EHDEN project is an effort to harmonize over 100 million health records to the OMOP Common Data Model (CDM) and creates the infrastructure for a European Health Data network for collaboration on real world healthcare data. EHDEN essentially aims to make healthcare data in Europe FAIR at an unprecedented level. The EHDEN framework contains a number of important applications to support this objective, and builds on an existing open source community: the Observational Health Data Sciences and Informatics (OHDSI, pronounced “Odyssey”) community. It is a multi-stakeholder, interdisciplinary collaborative to bring out the value of health data through large-scale analytics, which develops and maintains a number of key projects, such as the OMOP Common Data Model and Standardized Vocabularies.
The Hyve is a company focused on enabling open science by developing and implementing open source solutions and FAIRifying data in the life sciences [1]. The Hyve has a leadership role in the technical implementation (WP4) of the EHDEN project and also in the implementation of the FAIR principles in EHDEN.
Recently, the OHDSI community organized a so-called study-a-thon around COVID-19 [2,3] which was aimed at addressing a number of key open medical questions (e.g. real world safety of hydroxychloroquine, effectiveness of ACE inhibitors, prediction of hospitalization etc.) in a large collection of healthcare databases around the world. The EHDEN team played a key role in organizing this study-a-thon, and many important digital assets were produced, such as study protocols, database metadata and characterizations, study results, publications etc. In this use case, we describe a mechanism for making these digital assets FAIR in a real-time manner using widely adopted web standards such as JSON-LD.
Two key goals for this solution were:
- Provide visibility of the ongoing collaborative research to the rest of the world, both to humans and machines
- Create an overview and future reference for study-a-thon participants on the various digital assets created in the study process.
Process
FAIR Assessment: establishing the baseline
The FAIRification of study-a-thon data began with a FAIR assessment in order to better understand the current state of the data. The assessment covered two aspects: firstly, the studies themselves, including elements such as protocol and related publications, and secondly, the study’s data sources.
The FAIR assessment of the studies revealed that there was room for improvement in both Findability and Reusability, while the score was comparatively higher for Accessibility and Interoperability. Methods to improve Findability could include the implementation of an identifier scheme for both data and metadata, as well as using richer and more extensive metadata. Metadata, especially provenance metadata, would also increase the Reusability of the data. Data sources for a study are currently listed in a table, with a few columns of metadata for each entry. Here we saw improvement possibilities in Findability and Interoperability due to the lack of identifiers and structured data. Adding structure to the metadata, such as through external vocabularies, would greatly increase both the Interoperability and Findability of the data sources.
JSON-LD Schema.org: defining the semantic model
Since increasing Findability was a common denominator for both studies and data sources, we have been developing structured metadata elements tailored for studies and data sources. These elements are encoded using JSON-LD, which will be embedded in the study-a-thon website, thus creating a common definition of the data and metadata to both humans and machines. Representing the metadata in JSON-LD also increases the exposure of the study-a-thons to common search engines.
Metadata elements are based on schema.org, which is a broad and widely used vocabulary to make data on the web discoverable. The drawback of schema.org is that it is not extremely detailed, a compromise we settled on given the advantage of its popularity for web-based data. For each study and data source, a controlled set of metadata elements are chosen to describe an instance of each. For study, metadata covers concepts such as the type of study, the drug studied and information on the medical condition (Table 1). Similarly, for data sources, metadata is focused on the data source characteristics, such as the population covered in the data source, as well as provenance information. For a small percentage of the metadata, elements did not already exist in schema.org. In these cases, the vocabulary will be extended with custom concepts and relationships.
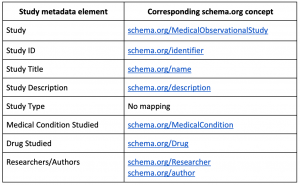
Table 1: An example of a few study metadata elements with the corresponding concept from schema.org. In the case of ‘Study Type’, which describes if the study is an estimation, prediction or characterization study, there is no equivalent concept in schema.org. ‘Study Type’ will map to a custom concept that will be encoded in the website’s JSON-LD, along with all the schema.org concepts.
Site generator: Publishing the digital assets for humans and machines
Once all the metadata for a study and data source has been defined and, where possible, mapped to its corresponding schema.org concept, these can be expressed in a machine-interoperable JSON-LD format to be implemented in the study-a-thon website. For this particular project, we chose a static site generator named Hugo, which already had a nice theme that we could leverage for this purpose [4]. The exact technology for the website is not very important with respect to the FAIR principles, but the advantage of choosing a static site generator is that it is very easy to take structured content (in this case expressed in YAML) and map that to both rendering to HTML and CSS for human readability as well as structured data rendering in the form of JSON-LD to enable machine readability. The Hugo project already has a site theme named academic that we adapted for this purpose [5].
Outcomes
This case study describes work in progress, so we may update it once we have finalized the approach and made it reusable for future study-a-thons and hackathons that involve real-time production of research assets such as study protocols, results, preprints etc. However, currently the result consists of a website that both highlights the key outcomes of the studyathon as human readable data, as well as exposes structured JSON-LD data to facilitate machine readability for search engines and web crawlers.
The end goal of EHDEN and also of this mini-project around the OHDSI COVID-19 studyathon is to create reusable frameworks for making (research in) healthcare data FAIR for the benefit of medical practice. This will ensure that once we have the next study-a-thon, there is an easy-to-use framework to quickly instantiate a website that covers both goals described in the introduction. Future work also needs to happen to make for example the generation of persistent identifiers easier, especially for auxiliary digital assets such as study protocols, cohort definitions, interactive study results etc. that are not scientific papers or pre-prints. Providing these services is the goal of projects such as EOSC.
References and Resources
- The Hyve https://thehyve.nl
- The Book of OHDSI – Open Science chapter: https://ohdsi.github.io/TheBookOfOhdsi/OpenScience.html
- OHDSI COVID-19 studyathon story: 88 hours
- Hugo static site generator: https://gohugo.io
- Hugo Theme named academic https://themes.gohugo.io/academic
At a Glance
Team(s)
- 2 FTE semantic modelling experts
- 1 FTE website developer
- 1 FTE OHDSI domain expert
Timing
- 3 weeks
Deliverables & benefits
- COVID-19 website (still under development)
- Semantic model expressed as subset of schema.org
- Reusable static site generator code

